Decomposition#
Decomposition techniques are most commonly used as a means of noise
reduction (or denoising) and dimensionality reduction. To apply a
decomposition to your dataset, run the decomposition()
method, for example:
>>> s = hs.signals.Signal1D(np.random.randn(10, 10, 200))
>>> s.decomposition()
Decomposition info:
normalize_poissonian_noise=False
algorithm=SVD
output_dimension=None
centre=None
>>> # Load data from a file, then decompose
>>> s = hs.load("my_file.hspy")
>>> s.decomposition()
Note
The signal s must be multi-dimensional, i.e.
s.axes_manager.navigation_size > 1
One of the most popular uses of decomposition()
is data denoising. This is achieved by using a limited set of components
to make a model of the original dataset, omitting the less significant components that
ideally contain only noise.
To reconstruct your denoised or reduced model, run the
get_decomposition_model() method. For example:
>>> # Use all components to reconstruct the model
>>> sc = s.get_decomposition_model()
>>> # Use first 3 components to reconstruct the model
>>> sc = s.get_decomposition_model(3)
>>> # Use components [0, 2] to reconstruct the model
>>> sc = s.get_decomposition_model([0, 2])
Sometimes, it is useful to examine the residuals between your original data and
the decomposition model. You can easily calculate and display the residuals,
since get_decomposition_model() returns a new
object, which in the example above we have called sc:
>>> (s - sc).plot()
You can perform operations on this new object sc later.
It is a copy of the original s object, except that the data has
been replaced by the model constructed using the chosen components.
If you provide the output_dimension argument, which takes an integer value,
the decomposition algorithm attempts to find the best approximation for the
dataset \(X\) with only a limited set of factors \(A\) and loadings \(B\),
such that \(X \approx A B^T\).
>>> s.decomposition(output_dimension=3)
Some of the algorithms described below require output_dimension to be provided.
Available algorithms#
HyperSpy implements a number of decomposition algorithms via the algorithm argument.
The table below lists the algorithms that are currently available, and includes
links to the appropriate documentation for more information on each one.
Note
Choosing which algorithm to use is likely to depend heavily on the nature of your dataset and the type of analysis you are trying to perform. We discuss some of the reasons for choosing one algorithm over another below, but would encourage you to do your own research as well. The scikit-learn documentation is a very good starting point.
Algorithm |
Method |
|---|---|
“SVD” (default) |
|
“MLPCA” |
|
“sklearn_pca” |
|
“NMF” |
|
“sparse_pca” |
|
“mini_batch_sparse_pca” |
|
“RPCA” |
|
“ORPCA” |
|
“ORNMF” |
|
custom object |
An object implementing |
Singular value decomposition (SVD)#
The default algorithm in HyperSpy is "SVD", which uses an approach called
“singular value decomposition” to decompose the data in the form
\(X = U \Sigma V^T\). The factors are given by \(U \Sigma\), and the
loadings are given by \(V^T\). For more information, please read the method
documentation for svd_pca().
>>> s = hs.signals.Signal1D(np.random.randn(10, 10, 200))
>>> s.decomposition()
Decomposition info:
normalize_poissonian_noise=False
algorithm=SVD
output_dimension=None
centre=None
Note
In some fields, including electron microscopy, this approach of applying an SVD directly to the data \(X\) is often called PCA (see below).
However, in the classical definition of PCA, the SVD should be applied to data that has first been “centered” by subtracting the mean, i.e. \(\mathrm{SVD}(X - \bar X)\).
The "SVD" algorithm in HyperSpy does not apply this
centering step by default. As a result, you may observe differences between
the output of the "SVD" algorithm and, for example,
sklearn.decomposition.PCA, which does apply centering.
Principal component analysis (PCA)#
One of the most popular decomposition methods is principal component analysis (PCA).
To perform PCA on your dataset, run the decomposition()
method with any of following arguments.
If you have scikit-learn installed:
>>> s.decomposition(algorithm="sklearn_pca")
Decomposition info:
normalize_poissonian_noise=False
algorithm=sklearn_pca
output_dimension=None
centre=None
scikit-learn estimator:
PCA()
You can also turn on centering with the default "SVD" algorithm via
the "centre" argument:
# Subtract the mean along the navigation axis
>>> s.decomposition(algorithm="SVD", centre="navigation")
Decomposition info:
normalize_poissonian_noise=False
algorithm=SVD
output_dimension=None
centre=navigation
# Subtract the mean along the signal axis
>>> s.decomposition(algorithm="SVD", centre="signal")
Decomposition info:
normalize_poissonian_noise=False
algorithm=SVD
output_dimension=None
centre=signal
You can also use sklearn.decomposition.PCA directly:
>>> from sklearn.decomposition import PCA
>>> s.decomposition(algorithm=PCA())
Decomposition info:
normalize_poissonian_noise=False
algorithm=PCA()
output_dimension=None
centre=None
scikit-learn estimator:
PCA()
Poissonian noise#
Most of the standard decomposition algorithms assume that the noise of the data follows a Gaussian distribution (also known as “homoskedastic noise”). In cases where your data is instead corrupted by Poisson noise, HyperSpy can “normalize” the data by performing a scaling operation, which can greatly enhance the result. More details about the normalization procedure can be found in [Keenan2004].
To apply Poissonian noise normalization to your data:
>>> s.decomposition(normalize_poissonian_noise=True)
>>> # Because it is the first argument we could have simply written:
>>> s.decomposition(True)
Warning
Poisson noise normalization cannot be used in combination with data
centering using the 'centre' argument. Attempting to do so will
raise an error.
Maximum likelihood principal component analysis (MLPCA)#
Instead of applying Poisson noise normalization to your data, you can instead use an approach known as Maximum Likelihood PCA (MLPCA), which provides a more robust statistical treatment of non-Gaussian “heteroskedastic noise”.
>>> s.decomposition(algorithm="MLPCA")
For more information, please read the method documentation for mlpca().
Note
You must set the output_dimension when using MLPCA.
Robust principal component analysis (RPCA)#
PCA is known to be very sensitive to the presence of outliers in data. These outliers can be the result of missing or dead pixels, X-ray spikes, or very low count data. If one assumes a dataset, \(X\), to consist of a low-rank component \(L\) corrupted by a sparse error component \(S\), such that \(X=L+S\), then Robust PCA (RPCA) can be used to recover the low-rank component for subsequent processing [Candes2011].
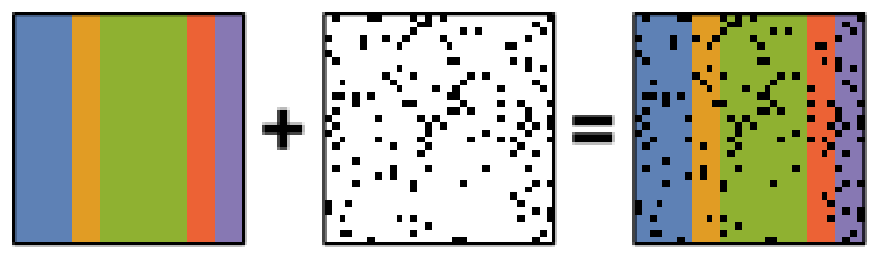
Schematic diagram of the robust PCA problem, which combines a low-rank matrix with sparse errors. Robust PCA aims to decompose the matrix back into these two components.#
Note
You must set the output_dimension when using Robust PCA.
The default RPCA algorithm is GoDec [Zhou2011]. In HyperSpy
it returns the factors and loadings of \(L\). RPCA solvers work by using
regularization, in a similar manner to lasso or ridge regression, to enforce
the low-rank constraint on the data. The low-rank regularization parameter,
lambda1, defaults to 1/sqrt(n_features), but it is strongly recommended
that you explore the behaviour of different values.
>>> s.decomposition(algorithm="RPCA", output_dimension=3, lambda1=0.1)
Decomposition info:
normalize_poissonian_noise=False
algorithm=RPCA
output_dimension=3
centre=None
HyperSpy also implements an online algorithm for RPCA developed by Feng et al. [Feng2013]. This minimizes memory usage, making it suitable for large datasets, and can often be faster than the default algorithm.
>>> s.decomposition(algorithm="ORPCA", output_dimension=3)
The online RPCA implementation sets several default parameters that are usually suitable for most datasets, including the regularization parameter highlighted above. Again, it is strongly recommended that you explore the behaviour of these parameters. To further improve the convergence, you can “train” the algorithm with the first few samples of your dataset. For example, the following code will train ORPCA using the first 32 samples of the data.
>>> s.decomposition(algorithm="ORPCA", output_dimension=3, training_samples=32)
Finally, online RPCA includes two alternatives methods to the default block-coordinate descent solver, which can again improve both the convergence and speed of the algorithm. These are particularly useful for very large datasets.
The methods are based on stochastic gradient descent (SGD), and take an additional parameter to set the learning rate. The learning rate dictates the size of the steps taken by the gradient descent algorithm, and setting it too large can lead to oscillations that prevent the algorithm from finding the correct minima. Usually a value between 1 and 2 works well:
>>> s.decomposition(algorithm="ORPCA",
... output_dimension=3,
... method="SGD",
... subspace_learning_rate=1.1)
You can also use Momentum Stochastic Gradient Descent (MomentumSGD),
which typically improves the convergence properties of stochastic gradient
descent. This takes the further parameter subspace_momentum, which should
be a fraction between 0 and 1.
>>> s.decomposition(algorithm="ORPCA",
... output_dimension=3,
... method="MomentumSGD",
... subspace_learning_rate=1.1,
... subspace_momentum=0.5)
Using the "SGD" or "MomentumSGD" methods enables the subspace,
i.e. the underlying low-rank component, to be tracked as it changes
with each sample update. The default method instead assumes a fixed,
static subspace.
Non-negative matrix factorization (NMF)#
Another popular decomposition method is non-negative matrix factorization (NMF), which can be accessed in HyperSpy with:
>>> s.decomposition(algorithm="NMF")
Unlike PCA, NMF forces the components to be strictly non-negative, which can aid the physical interpretation of components for count data such as images, EELS or EDS. For an example of NMF in EELS processing, see [Nicoletti2013].
NMF takes the optional argument output_dimension, which determines the number
of components to keep. Setting this to a small number is recommended to keep
the computation time small. Often it is useful to run a PCA decomposition first
and use the scree plot to determine a suitable value
for output_dimension.
Robust non-negative matrix factorization (RNMF)#
In a similar manner to the online, robust methods that complement PCA above, HyperSpy includes an online robust NMF method. This is based on the OPGD (Online Proximal Gradient Descent) algorithm of [Zhao2016].
Note
You must set the output_dimension when using Robust NMF.
As before, you can control the regularization applied via the parameter “lambda1”:
>>> s.decomposition(algorithm="ORNMF", output_dimension=3, lambda1=0.1)
The MomentumSGD method is useful for scenarios where the subspace, i.e. the underlying low-rank component, is changing over time.
>>> s.decomposition(algorithm="ORNMF",
... output_dimension=3,
... method="MomentumSGD",
... subspace_learning_rate=1.1,
... subspace_momentum=0.5)
Both the default and MomentumSGD solvers assume an l2-norm minimization problem, which can still be sensitive to very heavily corrupted data. A more robust alternative is available, although it is typically much slower.
>>> s.decomposition(algorithm="ORNMF", output_dimension=3, method="RobustPGD")
Custom decomposition algorithms#
HyperSpy supports passing a custom decomposition algorithm, provided it follows the form of a
scikit-learn estimator.
Any object that implements fit and transform methods is acceptable, including
sklearn.pipeline.Pipeline and sklearn.model_selection.GridSearchCV.
You can access the fitted estimator by passing return_info=True.
>>> # Passing a custom decomposition algorithm
>>> from sklearn.preprocessing import MinMaxScaler
>>> from sklearn.pipeline import Pipeline
>>> from sklearn.decomposition import PCA
>>> pipe = Pipeline([("scaler", MinMaxScaler()), ("PCA", PCA())])
>>> out = s.decomposition(algorithm=pipe, return_info=True)
Decomposition info:
normalize_poissonian_noise=False
algorithm=Pipeline(steps=[('scaler', MinMaxScaler()), ('PCA', PCA())])
output_dimension=None
centre=None
scikit-learn estimator:
Pipeline(steps=[('scaler', MinMaxScaler()), ('PCA', PCA())])
>>> out
Pipeline(steps=[('scaler', MinMaxScaler()), ('PCA', PCA())])