Electron Energy Loss Spectroscopy¶
Tools for EELS data analysis¶
The functions described in this chapter are only available for the
EELSSpectrum class. To transform a
BaseSignal (or subclass) into a
EELSSpectrum:
>>> s.set_signal_type("EELS")
Note these chapter discusses features that are available only for
EELSSpectrum class. However, this class inherits
many useful feature from its parent class that are documented in previous
chapters.
Elemental composition of the sample¶
It can be useful to define the elemental composition of the sample for
archiving purposes or to use some feature (e.g. curve fitting) that requieres
this information. The elemental composition of the sample can be declared
using add_elements(). The information is
stored in the metadata attribute (see
Metadata structure). This information is saved to file when saving in
the hdf5 format.
Thickness estimation¶
The estimate_thickness() can estimate the
thickness from a low-loss EELS spectrum using the Log-Ratio method.
Zero-loss peak centre and alignment¶
The estimate_zero_loss_peak_centre() can be used to estimate the position of the zero-loss peak. The method assumes that the ZLP is the most intense feature in the spectra. For a more general approach see find_peaks1D_ohaver().
The align_zero_loss_peak() can
align the ZLP with subpixel accuracy. It is more robust and easy to use than
align1D() for the task. Note that it is
possible to apply the same alignment to other spectra using the also_align
argument. This can be useful e.g. to align core-loss spectra acquired quasi-simultaneously.
If there are other features in the low loss signal which are more intense than the
ZLP, the signal_range argument can narrow down the energy range for searching for the
ZLP.
Deconvolutions¶
Three deconvolution methods are currently available:
Estimate elastic scattering intensity¶
The
estimate_elastic_scattering_intensity()
can be used to calculate the integral of the zero loss peak (elastic intensity)
from EELS low-loss spectra containing the zero loss peak using the
(rudimentary) threshold method. The threshold can be global or spectrum-wise.
If no threshold is provided it is automatically calculated using
estimate_elastic_scattering_threshold()
with default values.
estimate_elastic_scattering_threshold()
can be used to calculate separation point between elastic and inelastic
scattering on EELS low-loss spectra. This algorithm calculates the derivative
of the signal and assigns the inflexion point to the first point below a
certain tolerance. This tolerance value can be set using the tol keyword.
Currently, the method uses smoothing to reduce the impact of the noise in the
measure. The number of points used for the smoothing window can be specified by
the npoints keyword.
Kramers-Kronig Analysis¶
New in version 0.7.
The single-scattering EEL spectrum is approximately related to the complex
permittivity of the sample and can be estimated by Kramers-Kronig analysis.
The kramers_kronig_analysis() method
inplements the Kramers-Kronig FFT method as in [Egerton2011] to estimate the
complex dielectric funtion from a low-loss EELS spectrum. In addition, it can
estimate the thickness if the refractive index is known and approximately
correct for surface plasmon excitations in layers.
EELS curve fitting¶
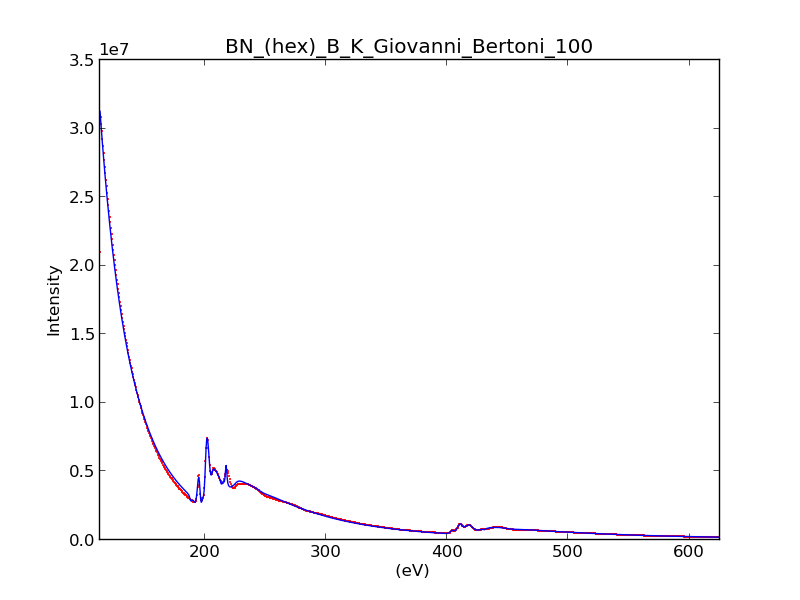
HyperSpy makes it really easy to quantify EELS core-loss spectra by curve fitting as it is shown in the next example of quantification of a boron nitride EELS spectrum from the The EELS Data Base.
Load the core-loss and low-loss spectra
>>> s = hs.load("BN_(hex)_B_K_Giovanni_Bertoni_100.msa")
>>> ll = hs.load("BN_(hex)_LowLoss_Giovanni_Bertoni_96.msa")
Set some important experimental information that is missing from the original core-loss file
>>> s.set_microscope_parameters(beam_energy=100, convergence_angle=0.2, collection_angle=2.55)
Warning
convergence_angle and collection_angle are actually semi-angles and are given in mrad. beam_energy is in keV.
Define the chemical composition of the sample
>>> s.add_elements(('B', 'N'))
In order to include the effect of plural scattering we provide the low-loss spectrum to create_model():
>>> m = s.create_model(ll=ll)
HyperSpy has created the model and configured it automatically:
>>> m.components
# | Attribute Name | Component Name | Component Type
---- | ------------------------- | ------------------------- | -------------------------
0 | PowerLaw | PowerLaw | PowerLaw
1 | N_K | N_K | EELSCLEdge
2 | B_K | B_K | EELSCLEdge
Conveniently, all the EELS core-loss components of the added elements are added automatically, names after its element symbol.
By default the fine structure features are disabled (although the default value can be configured (see Configuring HyperSpy). We must enable them to accurately fit this spectrum.
>>> m.enable_fine_structure()
We use smart_fit instead of standard fit method because smart_fit is optimized to fit EELS core-loss spectra
>>> m.smart_fit()
This fit can also be applied over the entire signal to fit a whole spectrum image
>>> m.multifit(kind='smart')
Print the result of the fit
>>> m.quantify()
Absolute quantification:
Elem. Intensity
B 0.045648
N 0.048061
Visualize the result
>>> m.plot()
There are several methods that are only available in
EELSModel:
smart_fit()is a fit method that is more robust than the standard routine when fitting EELS data.quantify()prints the intensity at the current locations of all the EELS ionisation edges in the model.remove_fine_structure_data()removes the fine structure spectral data range (as defined by thefine_structure_width)ionisation edge components. It is specially useful when fitting without convolving with a zero-loss peak.
The following methods permit to easily enable/disable background and ionisation edges components:
enable_edges()enable_background()disable_background()enable_fine_structure()disable_fine_structure()
The following methods permit to easily enable/disable several ionisation edge functionalities:
set_all_edges_intensities_positive()unset_all_edges_intensities_positive()enable_free_onset_energy()disable_free_onset_energy()fix_edges()free_edges()fix_fine_structure()free_fine_structure()
When fitting edges with fine structure enabled it is often desirable that the
fine structure region of nearby ionization edges does not overlap. HyperSpy
provides a method,
resolve_fine_structure(), to
automatically adjust the fine structure to prevent fine structure to avoid
overlapping. This method is executed automatically when e.g. components are
added or removed from the model, but sometimes is necessary to call it
manually.
New in version 0.7.1: Sometimes it is desirable to disable the automatic adjustment of the fine
structure width. It is possible to suspend this feature by calling
suspend_auto_fine_structure_width().
To resume it use
suspend_auto_fine_structure_width()