Note
Go to the end to download the full example code.
Simple simulation (2 Gaussians)#
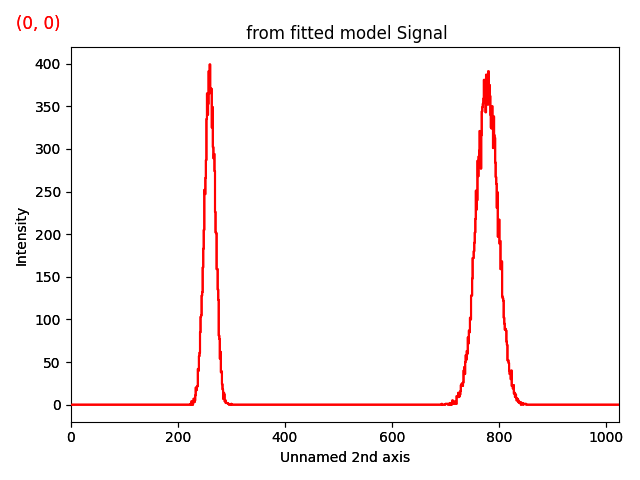
Creates a 2D hyperspectrum consisting of two Gaussians and plots it.
This example can serve as starting point to test other functionalities on the simulated hyperspectrum.
0%| | 0/1024 [00:00<?, ?it/s]
import numpy as np
import hyperspy.api as hs
import matplotlib.pyplot as plt
# Create an empty spectrum
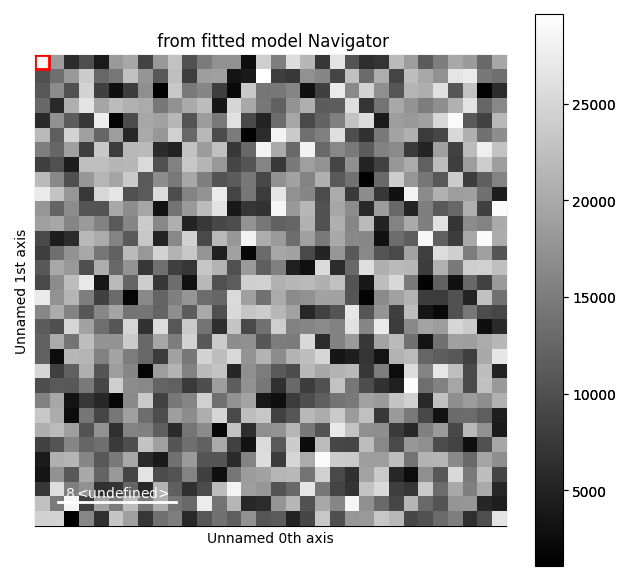
s = hs.signals.Signal1D(np.zeros((32, 32, 1024)))
# Generate some simple data: two Gaussians with random centers and area
# First we create a model
m = s.create_model()
# Define the first gaussian
gs1 = hs.model.components1D.Gaussian()
# Add it to the model
m.append(gs1)
# Set the parameters
gs1.sigma.value = 10
# Make the center vary in the -5,5 range around 128
gs1.centre.map['values'][:] = 256 + (np.random.random((32, 32)) - 0.5) * 10
gs1.centre.map['is_set'][:] = True
# Make the area vary between 0 and 10000
gs1.A.map['values'][:] = 10000 * np.random.random((32, 32))
gs1.A.map['is_set'][:] = True
# Second gaussian
gs2 = hs.model.components1D.Gaussian()
# Add it to the model
m.append(gs2)
# Set the parameters
gs2.sigma.value = 20
# Make the center vary in the -10,10 range around 768
gs2.centre.map['values'][:] = 768 + (np.random.random((32, 32)) - 0.5) * 20
gs2.centre.map['is_set'][:] = True
# Make the area vary between 0 and 20000
gs2.A.map['values'][:] = 20000 * np.random.random((32, 32))
gs2.A.map['is_set'][:] = True
# Create the dataset
s_model = m.as_signal()
# Add noise
s_model.set_signal_origin("simulation")
s_model.add_poissonian_noise()
# Plot the result
s_model.plot()
plt.show()
Total running time of the script: (0 minutes 1.274 seconds)