Model fitting
HyperSpy can perform curve fitting of one-dimensional signals (spectra) and
two-dimensional signals (images) in n-dimensional data sets.
Models are defined by adding individual functions (components in HyperSpy’s
terminology) to a BaseModel instance. Those individual
components are then summed to create the final model function that can be
fitted to the data, by adjusting the free parameters of the individual
components.
Models can be created and fit to experimental data in both one and two
dimensions i.e. spectra and images respectively. Most of the syntax is
identical in either case. A one-dimensional model is created when a model
is created for a Signal1D whereas a two-
dimensional model is created for a Signal2D.
Note
Plotting and analytical gradient-based fitting methods are not yet
implemented for the Model2D class.
Caveats
Before creating a model verify that the
is_binnedattribute of the signal axis is set to the correct value because the resulting model depends on this parameter. See Binned and unbinned signals for more details.When importing data that has been binned using other software, in particular Gatan’s DM, the stored values may be the averages of the binned channels or pixels, instead of their sum, as would be required for proper statistical analysis. We therefore cannot guarantee that the statistics will be valid, and so strongly recommend that all pre-fitting binning is performed using Hyperspy.
Creating a model
A Model1D can be created for data in the
Signal1D class using the
create_model() method:
>>> s = hs.signals.Signal1D(np.arange(300).reshape(30, 10))
>>> m = s.create_model() # Creates the 1D-Model and assign it to m
Similarly, a Model2D can be created for data
in the Signal2D class using the
create_model() method:
>>> im = hs.signals.Signal2D(np.arange(300).reshape(3, 10, 10))
>>> mod = im.create_model() # Create the 2D-Model and assign it to mod
The syntax for creating both one-dimensional and two-dimensional models is thus
identical for the user in practice. When a model is created you may be
prompted to provide important information not already included in the
datafile, e.g. if s is EELS data, you may be asked for the accelerating
voltage, convergence and collection semi-angles etc.
Model components
In HyperSpy a model consists of a sum of individual components. For convenience, HyperSpy provides a number of pre-defined model components as well as mechanisms to create your own components.
Pre-defined model components
Various components are available in one (components1d) and
two-dimensions (components2d) to construct a
model.
The following general components are currently available for one-dimensional models:
The following components developed with specific signal types in mind are currently available for one-dimensional models:
The following components are currently available for two-dimensional models:
However, this doesn’t mean that you have to limit yourself to this meagre list of functions. As discussed below, it is very easy to turn a mathematical, fixed-pattern or Python function into a component.
Define components from a mathematical expression
The easiest way to turn a mathematical expression into a component is using the
Expression component. For example, the
following is all you need to create a
Gaussian component with more sensible
parameters for spectroscopy than the one that ships with HyperSpy:
>>> g = hs.model.components1D.Expression(
... expression="height * exp(-(x - x0) ** 2 * 4 * log(2)/ fwhm ** 2)",
... name="Gaussian",
... position="x0",
... height=1,
... fwhm=1,
... x0=0,
... module="numpy")
If the expression is inconvenient to write out in full (e.g. it’s long and/or complicated), multiple substitutions can be given, separated by semicolons. Both symbolic and numerical substitutions are allowed:
>>> expression = "h / sqrt(p2) ; p2 = 2 * m0 * e1 * x * brackets;"
>>> expression += "brackets = 1 + (e1 * x) / (2 * m0 * c * c) ;"
>>> expression += "m0 = 9.1e-31 ; c = 3e8; e1 = 1.6e-19 ; h = 6.6e-34"
>>> wavelength = hs.model.components1D.Expression(
... expression=expression,
... name="Electron wavelength with voltage")
Expression uses Sympy internally to turn the string into
a function. By default it “translates” the expression using
numpy, but often it is possible to boost performance by using
numexpr instead.
It can also create 2D components with optional rotation. In the following example we create a 2D Gaussian that rotates around its center:
>>> g = hs.model.components2D.Expression(
... "k * exp(-((x-x0)**2 / (2 * sx ** 2) + (y-y0)**2 / (2 * sy ** 2)))",
... "Gaussian2d", add_rotation=True, position=("x0", "y0"),
... module="numpy", )
Define new components from a Python function
Of course Expression is only useful for
analytical functions. You can define more general components modifying the
following template to suit your needs:
from hyperspy.component import Component
class MyComponent(Component):
"""
"""
def __init__(self, parameter_1=1, parameter_2=2):
# Define the parameters
Component.__init__(self, ('parameter_1', 'parameter_2'))
# Optionally we can set the initial values
self.parameter_1.value = parameter_1
self.parameter_2.value = parameter_2
# The units (optional)
self.parameter_1.units = 'Tesla'
self.parameter_2.units = 'Kociak'
# Once defined we can give default values to the attribute
# For example we fix the attribure_1 (optional)
self.parameter_1.attribute_1.free = False
# And we set the boundaries (optional)
self.parameter_1.bmin = 0.
self.parameter_1.bmax = None
# Optionally, to boost the optimization speed we can also define
# the gradients of the function we the syntax:
# self.parameter.grad = function
self.parameter_1.grad = self.grad_parameter_1
self.parameter_2.grad = self.grad_parameter_2
# Define the function as a function of the already defined parameters,
# x being the independent variable value
def function(self, x):
p1 = self.parameter_1.value
p2 = self.parameter_2.value
return p1 + x * p2
# Optionally define the gradients of each parameter
def grad_parameter_1(self, x):
"""
Returns d(function)/d(parameter_1)
"""
return 0
def grad_parameter_2(self, x):
"""
Returns d(function)/d(parameter_2)
"""
return x
Define components from a fixed-pattern
The ScalableFixedPattern
component enables fitting a pattern (in the form of a
Signal1D instance) to data by shifting
(shift)
and
scaling it in the x and y directions using the
xscale
and
yscale
parameters respectively.
Adding components to the model
To print the current components in a model use
components. A table with component number,
attribute name, component name and component type will be printed:
>>> m
<Model, title: my signal title>
>>> m.components # an empty model
# | Attribute Name | Component Name | Component Type
---- | -------------------- | -------------------- | ---------------------
Note
Sometimes components may be created automatically. For example, if
the Signal1D is recognised as EELS data, a
power-law background component may automatically be added to the model.
Therefore, the table above may not all may empty on model creation.
To add a component to the model, first we have to create an instance of the
component.
Once the instance has been created we can add the component to the model
using the append() and
extend() methods for one or more components
respectively.
As an example, let’s add several Gaussian
components to the model:
>>> gaussian = hs.model.components1D.Gaussian() # Create a Gaussian comp.
>>> m.append(gaussian) # Add it to the model
>>> m.components # Print the model components
# | Attribute Name | Component Name | Component Type
---- | -------------------- | --------------------- | ---------------------
0 | Gaussian | Gaussian | Gaussian
>>> gaussian2 = hs.model.components1D.Gaussian() # Create another gaussian
>>> gaussian3 = hs.model.components1D.Gaussian() # Create a third gaussian
We could use the append() method twice to add the
two Gaussians, but when adding multiple components it is handier to use the
extend method that enables adding a list of components at once.
>>> m.extend((gaussian2, gaussian3)) # note the double parentheses!
>>> m.components
# | Attribute Name | Component Name | Component Type
---- | -------------------- | ------------------- | ---------------------
0 | Gaussian | Gaussian | Gaussian
1 | Gaussian_0 | Gaussian_0 | Gaussian
2 | Gaussian_1 | Gaussian_1 | Gaussian
We can customise the name of the components.
>>> gaussian.name = 'Carbon'
>>> gaussian2.name = 'Long Hydrogen name'
>>> gaussian3.name = 'Nitrogen'
>>> m.components
# | Attribute Name | Component Name | Component Type
---- | --------------------- | --------------------- | -------------------
0 | Carbon | Carbon | Gaussian
1 | Long_Hydrogen_name | Long Hydrogen name | Gaussian
2 | Nitrogen | Nitrogen | Gaussian
Notice that two components cannot have the same name:
>>> gaussian2.name = 'Carbon'
Traceback (most recent call last):
File "<ipython-input-5-2b5669fae54a>", line 1, in <module>
g2.name = "Carbon"
File "/home/fjd29/Python/hyperspy/hyperspy/component.py", line 466, in
name "the name " + str(value))
ValueError: Another component already has the name Carbon
It is possible to access the components in the model by their name or by the index in the model.
>>> m
# | Attribute Name | Component Name | Component Type
---- | --------------------- | -------------------- | -------------------
0 | Carbon | Carbon | Gaussian
1 | Long_Hydrogen_name | Long Hydrogen name | Gaussian
2 | Nitrogen | Nitrogen | Gaussian
>>> m[0]
<Carbon (Gaussian component)>
>>> m["Long Hydrogen name"]
<Long Hydrogen name (Gaussian component)>
In addition, the components can be accessed in the
components Model attribute. This is specially
useful when working in interactive data analysis with IPython because it
enables tab completion.
>>> m
# | Attribute Name | Component Name | Component Type
---- | --------------------- | --------------------- | -------------------
0 | Carbon | Carbon | Gaussian
1 | Long_Hydrogen_name | Long Hydrogen name | Gaussian
2 | Nitrogen | Nitrogen | Gaussian
>>> m.components.Long_Hydrogen_name
<Long Hydrogen name (Gaussian component)>
It is possible to “switch off” a component by setting its
active attribute to False. When a component is
switched off, to all effects it is as if it was not part of the model. To
switch it back on simply set the active attribute back to True.
In multi-dimensional signals it is possible to store the value of the
active attribute at each navigation index.
To enable this feature for a given component set the
active_is_multidimensional attribute to
True.
>>> s = hs.signals.Signal1D(np.arange(100).reshape(10,10))
>>> m = s.create_model()
>>> g1 = hs.model.components1D.Gaussian()
>>> g2 = hs.model.components1D.Gaussian()
>>> m.extend([g1,g2])
>>> g1.active_is_multidimensional = True
>>> g1._active_array
array([ True, True, True, True, True, True, True, True, True, True], dtype=bool)
>>> g2._active_array is None
True
>>> m.set_component_active_value(False)
>>> g1._active_array
array([False, False, False, False, False, False, False, False, False, False], dtype=bool)
>>> m.set_component_active_value(True, only_current=True)
>>> g1._active_array
array([ True, False, False, False, False, False, False, False, False, False], dtype=bool)
>>> g1.active_is_multidimensional = False
>>> g1._active_array is None
True
Indexing the model
Often it is useful to consider only part of the model - for example at
a particular location (i.e. a slice in the navigation space) or energy range
(i.e. a slice in the signal space). This can be done using exactly the same
syntax that we use for signal indexing (Indexing).
red_chisq and dof
are automatically recomputed for the resulting slices.
>>> s = hs.signals.Signal1D(np.arange(100).reshape(10,10))
>>> m = s.create_model()
>>> m.append(hs.model.components1D.Gaussian())
>>> # select first three navigation pixels and last five signal channels
>>> m1 = m.inav[:3].isig[-5:]
>>> m1.signal
<Signal1D, title: , dimensions: (3|5)>
Getting and setting parameter values and attributes
print_current_values() prints the properties of the
parameters of the components in the current coordinates. In the Jupyter Notebook,
the default view is HTML-formatted, which allows for formatted copying
into other software, such as Excel. This can be changed to a standard
terminal view with the argument fancy=False. One can also filter for only active
components and only showing component with free parameters with the arguments
only_active and only_free, respectively.
The current values of a particular component can be printed using the
print_current_values() method.
>>> m = s.create_model()
>>> m.fit()
>>> G = m[1]
>>> G.print_current_values(fancy=False)
Gaussian: Al_Ka
Active: True
Parameter Name | Free | Value | Std | Min
============== | ===== | ========== | ========== | ==========
A | True | 62894.6824 | 1039.40944 | 0.0
sigma | False | 0.03253440 | None | None
centre | False | 1.4865 | None | None
The current coordinates can be either set by navigating the
plot(), or specified by pixel indices in
m.axes_manager.indices or as calibrated coordinates in
m.axes_manager.coordinates.
parameters contains a list of the parameters
of a component and free_parameters lists only
the free parameters.
The value of a particular parameter in the current coordinates can be
accessed by component.Parameter.value (e.g. Gaussian.A.value).
To access an array of the value of the parameter across all navigation
pixels, component.Parameter.map['values'] (e.g.
Gaussian.A.map["values"]) can be used. On its own,
component.Parameter.map returns a NumPy array with three elements:
'values', 'std' and 'is_set'. The first two give the value and
standard error for each index. The last element shows whether the value has
been set in a given index, either by a fitting procedure or manually.
If a model contains several components with the same parameters, it is possible
to change them all by using set_parameters_value().
Example:
>>> s = hs.signals.Signal1D(np.arange(100).reshape(10,10))
>>> m = s.create_model()
>>> g1 = hs.model.components1D.Gaussian()
>>> g2 = hs.model.components1D.Gaussian()
>>> m.extend([g1,g2])
>>> m.set_parameters_value('A', 20)
>>> g1.A.map['values']
array([ 20., 20., 20., 20., 20., 20., 20., 20., 20., 20.])
>>> g2.A.map['values']
array([ 20., 20., 20., 20., 20., 20., 20., 20., 20., 20.])
>>> m.set_parameters_value('A', 40, only_current=True)
>>> g1.A.map['values']
array([ 40., 20., 20., 20., 20., 20., 20., 20., 20., 20.])
>>> m.set_parameters_value('A',30, component_list=[g2])
>>> g2.A.map['values']
array([ 30., 30., 30., 30., 30., 30., 30., 30., 30., 30.])
>>> g1.A.map['values']
array([ 40., 20., 20., 20., 20., 20., 20., 20., 20., 20.])
To set the free state of a parameter change the
free attribute. To change the free state
of all parameters in a component to True use
set_parameters_free(), and
set_parameters_not_free() for setting them to
False. Specific parameter-names can also be specified by using
parameter_name_list, shown in the example:
>>> g = hs.model.components1D.Gaussian()
>>> g.free_parameters
[<Parameter A of Gaussian component>,
<Parameter sigma of Gaussian component>,
<Parameter centre of Gaussian component>]
>>> g.set_parameters_not_free()
>>> g.set_parameters_free(parameter_name_list=['A','centre'])
>>> g.free_parameters
[<Parameter A of Gaussian component>,
<Parameter centre of Gaussian component>]
Similar functions exist for BaseModel:
set_parameters_free() and
set_parameters_not_free(). Which sets the
free states for the parameters in components in a model. Specific
components and parameter-names can also be specified. For example:
>>> g1 = hs.model.components1D.Gaussian()
>>> g2 = hs.model.components1D.Gaussian()
>>> m.extend([g1,g2])
>>> m.set_parameters_not_free()
>>> g1.free_parameters
[]
>>> g2.free_parameters
[]
>>> m.set_parameters_free(parameter_name_list=['A'])
>>> g1.free_parameters
[<Parameter A of Gaussian component>]
>>> g2.free_parameters
[<Parameter A of Gaussian component>]
>>> m.set_parameters_free([g1], parameter_name_list=['sigma'])
>>> g1.free_parameters
[<Parameter A of Gaussian component>,
<Parameter sigma of Gaussian component>]
>>> g2.free_parameters
[<Parameter A of Gaussian component>]
The value of a parameter can be coupled to the value of another by setting the
twin attribute:
>>> gaussian.parameters # Print the parameters of the Gaussian components
(<Parameter A of Carbon component>,
<Parameter sigma of Carbon component>,
<Parameter centre of Carbon component>)
>>> gaussian.centre.free = False # Fix the centre
>>> gaussian.free_parameters # Print the free parameters
[<Parameter A of Carbon component>, <Parameter sigma of Carbon component>]
>>> m.print_current_values(only_free=True, fancy=False) # Print the values of all free parameters.
Model1D:
Gaussian: Carbon
Active: True
Parameter Name | Free | Value | Std | Min | Max
============== | ===== | ========== | ========== | ========== | ==========
A | True | 1.0 | None | 0.0 | None
sigma | True | 1.0 | None | None | None
Gaussian: Long Hydrogen name
Active: True
Parameter Name | Free | Value | Std | Min | Max
============== | ===== | ========== | ========== | ========== | ==========
A | True | 1.0 | None | 0.0 | None
sigma | True | 1.0 | None | None | None
centre | True | 0.0 | None | None | None
Gaussian: Nitrogen
Active: True
Parameter Name | Free | Value | Std | Min | Max
============== | ===== | ========== | ========== | ========== | ==========
A | True | 1.0 | None | 0.0 | None
sigma | True | 1.0 | None | None | None
centre | True | 0.0 | None | None | None
>>> # Couple the A parameter of gaussian2 to the A parameter of gaussian 3:
>>> gaussian2.A.twin = gaussian3.A
>>> gaussian2.A.value = 10 # Set the gaussian2 A value to 10
>>> gaussian3.print_current_values(fancy=False)
Gaussian: Nitrogen
Active: True
Parameter Name | Free | Value | Std | Min | Max
============== | ===== | ========== | ========== | ========== | ==========
A | True | 10.0 | None | 0.0 | None
sigma | True | 1.0 | None | None | None
centre | True | 0.0 | None | None | None
>>> gaussian3.A.value = 5 # Set the gaussian1 centre value to 5
>>> gaussian2.print_current_values(fancy=False)
Gaussian: Long Hydrogen name
Active: True
Parameter Name | Free | Value | Std | Min | Max
============== | ===== | ========== | ========== | ========== | ==========
A | False | 5.0 | None | 0.0 | None
sigma | True | 1.0 | None | None | None
centre | True | 0.0 | None | None | None
Deprecated since version 1.2.0: Setting the twin_function and
twin_inverse_function attributes. Set the
twin_function_expr and
twin_inverse_function_expr attributes
instead.
New in version 1.2.0: twin_function_expr and
twin_inverse_function_expr.
By default the coupling function is the identity function. However it is
possible to set a different coupling function by setting the
twin_function_expr and
twin_inverse_function_expr attributes. For
example:
>>> gaussian2.A.twin_function_expr = "x**2"
>>> gaussian2.A.twin_inverse_function_expr = "sqrt(abs(x))"
>>> gaussian2.A.value = 4
>>> gaussian3.print_current_values(fancy=False)
Gaussian: Nitrogen
Active: True
Parameter Name | Free | Value | Std | Min | Max
============== | ===== | ========== | ========== | ========== | ==========
A | True | 2.0 | None | 0.0 | None
sigma | True | 1.0 | None | None | None
centre | True | 0.0 | None | None | None
>>> gaussian3.A.value = 4
>>> gaussian2.print_current_values(fancy=False)
Gaussian: Long Hydrogen name
Active: True
Parameter Name | Free | Value | Std | Min | Max
============== | ===== | ========== | ========== | ========== | ==========
A | False | 16.0 | None | 0.0 | None
sigma | True | 1.0 | None | None | None
centre | True | 0.0 | None | None | None
Fitting the model to the data
To fit the model to the data at the current coordinates (e.g. to fit one
spectrum at a particular point in a spectrum-image), use
fit(). HyperSpy implements a number of
different optimization approaches, each of which can have particular
benefits and/or drawbacks depending on your specific application.
A good approach to choosing an optimization approach is to ask yourself
the question “Do you want to…”:
Apply bounds to your model parameter values?
Use gradient-based fitting algorithms to accelerate your fit?
Estimate the standard deviations of the parameter values found by the fit?
Fit your data in the least-squares sense, or use another loss function?
Find the global optima for your parameters, or is a local optima acceptable?
Optimization algorithms
The following table summarizes the features of some of the optimizers currently available in HyperSpy, including whether they support parameter bounds, gradients and parameter error estimation. The “Type” column indicates whether the optimizers find a local or global optima.
Optimizer |
Bounds |
Gradients |
Errors |
Loss function |
Type |
Linear |
|---|---|---|---|---|---|---|
|
Yes |
Yes |
Yes |
Only |
local |
No |
|
Yes |
Yes |
Yes |
Only |
local |
No |
|
Yes |
Yes |
Yes |
Only |
local |
No |
|
No |
Yes |
Yes |
Only |
local |
No |
|
No |
No |
Yes [1] |
Only |
global |
Yes |
|
No |
No |
Yes [1] |
Only |
global |
Yes |
Yes [2] |
Yes [2] |
No |
All |
local |
No |
|
|
Yes |
No |
No |
All |
global |
No |
|
Yes |
No |
No |
All |
global |
No |
|
Yes |
No |
No |
All |
global |
No |
Footnotes
The default optimizer in HyperSpy is "lm", which stands for the Levenberg-Marquardt
algorithm. In
earlier versions of HyperSpy (< 1.6) this was known as "leastsq".
Loss functions
HyperSpy supports a number of loss functions. The default is "ls",
i.e. the least-squares loss. For the vast majority of cases, this loss
function is appropriate, and has the additional benefit of supporting
parameter error estimation and goodness-of-fit
testing. However, if your data contains very low counts per pixel, or
is corrupted by outliers, the "ML-poisson" and "huber" loss
functions may be worth investigating.
Least squares with error estimation
The following example shows how to perfom least squares optimization with
error estimation. First we create data consisting of a line
y = a*x + b with a = 1 and b = 100, and we then add Gaussian
noise to it:
>>> s = hs.signals.Signal1D(np.arange(100, 300))
>>> s.add_gaussian_noise(std=100)
To fit it, we create a model consisting of a
Polynomial component of order 1 and fit it
to the data.
>>> m = s.create_model()
>>> line = hs.model.components1D.Polynomial(order=1)
>>> m.append(line)
>>> m.fit()
Once the fit is complete, the optimized value of the parameters and their estimated standard deviation are stored in the following line attributes:
>>> line.a.value
0.9924615648843765
>>> line.b.value
103.67507406125888
>>> line.a.std
0.11771053738516088
>>> line.b.std
13.541061301257537
Warning
When the noise is heteroscedastic, only if the
metadata.Signal.Noise_properties.variance attribute of the
Signal1D instance is defined can
the parameter standard deviations be estimated accurately.
If the variance is not defined, the standard deviations are still computed, by setting variance equal to 1. However, this calculation will not be correct unless an accurate value of the variance is provided. See Setting the noise properties for more information.
Weighted least squares with error estimation
In the following example, we add Poisson noise to the data instead of Gaussian noise, and proceed to fit as in the previous example.
>>> s = hs.signals.Signal1D(np.arange(300))
>>> s.add_poissonian_noise()
>>> m = s.create_model()
>>> line = hs.model.components1D.Polynomial(order=1)
>>> m.append(line)
>>> m.fit()
>>> line.coefficients.value
(1.0052331707848698, -1.0723588390873573)
>>> line.coefficients.std
(0.0081710549764721901, 1.4117294994070277)
Because the noise is heteroscedastic, the least squares optimizer estimation is biased. A more accurate result can be obtained with weighted least squares, where the weights are proportional to the inverse of the noise variance. Although this is still biased for Poisson noise, it is a good approximation in most cases where there are a sufficient number of counts per pixel.
>>> exp_val = hs.signals.Signal1D(np.arange(300))
>>> s.estimate_poissonian_noise_variance(expected_value=exp_val)
>>> m.fit()
>>> line.coefficients.value
(1.0004224896604759, -0.46982916592391377)
>>> line.coefficients.std
(0.0055752036447948173, 0.46950832982673557)
Warning
When the attribute metadata.Signal.Noise_properties.variance
is defined, the behaviour is to perform a weighted least-squares
fit using the inverse of the noise variance as the weights.
In this scenario, to then disable weighting, you will need to unset
the attribute. You can achieve this with
set_noise_variance():
>>> m.signal.set_noise_variance(None)
>>> m.fit() # This will now be an unweighted fit
>>> line.coefficients.value
(1.0052331707848698, -1.0723588390873573)
Poisson maximum likelihood estimation
To avoid biased estimation in the case of data corrupted by Poisson noise with very few counts, we can use Poisson maximum likelihood estimation (MLE) instead. This is an unbiased estimator for Poisson noise. To perform MLE, we must use a general, non-linear optimizer from the table above, such as Nelder-Mead or L-BFGS-B:
>>> m.fit(optimizer="Nelder-Mead", loss_function="ML-poisson")
>>> line.coefficients.value
(1.0030718094185611, -0.63590210946134107)
Estimation of the parameter errors is not currently supported for Poisson maximum likelihood estimation.
Huber loss function
HyperSpy also implements the Huber loss function, which is typically less sensitive to outliers in the data compared to the least-squares loss. Again, we need to use one of the general non-linear optimization algorithms:
>>> m.fit(optimizer="Nelder-Mead", loss_function="huber")
Estimation of the parameter errors is not currently supported for the Huber loss function.
Custom loss functions
As well as the built-in loss functions described above, a custom loss function can be passed to the model:
>>> def my_custom_function(model, values, data, weights=None):
... """
... Parameters
... ----------
... model : Model instance
... the model that is fitted.
... values : np.ndarray
... A one-dimensional array with free parameter values suggested by the
... optimizer (that are not yet stored in the model).
... data : np.ndarray
... A one-dimensional array with current data that is being fitted.
... weights : {np.ndarray, None}
... An optional one-dimensional array with parameter weights.
...
... Returns
... -------
... score : float
... A signle float value, representing a score of the fit, with
... lower values corresponding to better fits.
... """
... # Almost any operation can be performed, for example:
... # First we store the suggested values in the model
... model.fetch_values_from_array(values)
...
... # Evaluate the current model
... cur_value = model(onlyactive=True)
...
... # Calculate the weighted difference with data
... if weights is None:
... weights = 1
... difference = (data - cur_value) * weights
...
... # Return squared and summed weighted difference
... return (difference**2).sum()
>>> # We must use a general non-linear optimizer
>>> m.fit(optimizer='Nelder-Mead', loss_function=my_custom_function)
If the optimizer requires an analytical gradient function, it can be similarly passed, using the following signature:
>>> def my_custom_gradient_function(model, values, data, weights=None):
... """
... Parameters
... ----------
... model : Model instance
... the model that is fitted.
... values : np.ndarray
... A one-dimensional array with free parameter values suggested by the
... optimizer (that are not yet stored in the model).
... data : np.ndarray
... A one-dimensional array with current data that is being fitted.
... weights : {np.ndarray, None}
... An optional one-dimensional array with parameter weights.
...
... Returns
... -------
... gradients : np.ndarray
... a one-dimensional array of gradients, the size of `values`,
... containing each parameter gradient with the given values
... """
... # As an example, estimate maximum likelihood gradient:
... model.fetch_values_from_array(values)
... cur_value = model(onlyactive=True)
...
... # We use in-built jacobian estimation
... jac = model._jacobian(values, data)
...
... return -(jac * (data / cur_value - 1)).sum(1)
>>> # We must use a general non-linear optimizer again
>>> m.fit(optimizer='L-BFGS-B',
... loss_function=my_custom_function,
... grad=my_custom_gradient_function)
Using gradient information
New in version 1.6: grad="analytical" and grad="fd" keyword arguments
Optimization algorithms that take into account the gradient of the loss function will often out-perform so-called “derivative-free” optimization algorithms in terms of how rapidly they converge to a solution. HyperSpy can use analytical gradients for model-fitting, as well as numerical estimates of the gradient based on finite differences.
If all the components in the model support analytical gradients,
you can pass grad="analytical" in order to use this information
when fitting. The results are typically more accurate than an
estimated gradient, and the optimization often runs faster since
fewer function evaluations are required to calculate the gradient.
Following the above examples:
>>> m = s.create_model()
>>> line = hs.model.components1D.Polynomial(order=1)
>>> m.append(line)
>>> # Use a 2-point finite-difference scheme to estimate the gradient
>>> m.fit(grad="fd", fd_scheme="2-point")
>>> # Use the analytical gradient
>>> m.fit(grad="analytical")
>>> # Huber loss and Poisson MLE functions
>>> # also support analytical gradients
>>> m.fit(grad="analytical", loss_function="huber")
>>> m.fit(grad="analytical", loss_function="ML-poisson")
Note
Analytical gradients are not yet implemented for the
Model2D class.
Bounded optimization
Non-linear optimization can sometimes fail to converge to a good optimum, especially if poor starting values are provided. Problems of ill-conditioning and non-convergence can be improved by using bounded optimization.
All components’ parameters have the attributes parameter.bmin and
parameter.bmax (“bounded min” and “bounded max”). When fitting using the
bounded=True argument by m.fit(bounded=True) or m.multifit(bounded=True),
these attributes set the minimum and maximum values allowed for parameter.value.
Currently, not all optimizers support bounds - see the
table above. In the following example, a Gaussian
histogram is fitted using a Gaussian
component using the Levenberg-Marquardt (“lm”) optimizer and bounds
on the centre parameter.
>>> s = hs.signals.BaseSignal(np.random.normal(loc=10, scale=0.01,
... size=100000)).get_histogram()
>>> s.axes_manager[-1].is_binned = True
>>> m = s.create_model()
>>> g1 = hs.model.components1D.Gaussian()
>>> m.append(g1)
>>> g1.centre.value = 7
>>> g1.centre.bmin = 7
>>> g1.centre.bmax = 14
>>> m.fit(optimizer="lm", bounded=True)
>>> m.print_current_values(fancy=False)
Model1D: histogram
Gaussian: Gaussian
Active: True
Parameter Name | Free | Value | Std | Min | Max
============== | ===== | ========== | ========== | ========== | ==========
A | True | 99997.3481 | 232.333693 | 0.0 | None
sigma | True | 0.00999184 | 2.68064163 | None | None
centre | True | 9.99980788 | 2.68064070 | 7.0 | 14.0
Linear least squares
New in version 1.7.
Linear fitting can be used to address some of the drawbacks of non-linear optimization:
it doesn’t suffer from the starting parameters issue, which can sometimes be problematic with nonlinear fitting. Since linear fitting uses linear algebra to find the solution (find the parameter values of the model), the solution is a unique solution, while nonlinear optimization uses an iterative approach and therefore relies on the initial values of the parameters.
it is fast, because i) in favorable situations, the signal can be fitted in a vectorized fashion, i.e. the signal is fitted in a single run instead of iterating over the navigation dimension; ii) it is not iterative, i.e. it does the calculation only one time instead of 10-100 iterations, depending on how quickly the non-linear optimizer will converge.
However, linear fitting can only fit linear models and will not be able to fit parameters which vary non-linearly.
A component is considered linear when its free parameters scale the component only
in the y-axis. For the exemplary function y = a*x**b, a is a linear parameter, whilst b
is not. If b.free = False, then the component is linear.
Components can also be made up of several linear parts. For instance,
the 2D-polynomial y = a*x**2+b*y**2+c*x+d*y+e is entirely linear.
Note
After creating a model with values for the nonlinear parameters, a quick way to set
all nonlinear parameters to be free = False is to use m.set_parameters_not_free(only_nonlinear=True)
To check if a parameter is linear, use the model or component method
print_current_values(). For a component to be
considered linear, it can hold only one free parameter, and that parameter
must be linear.
If all components in a model are linear, then a linear optimizer can be used to solve the problem as a linear regression problem! This can be done using two approaches:
the standard pixel-by-pixel approach as used by the nonlinear optimizers
fit the entire dataset in one vectorised operation, which will be much faster (up to 1000 times). However, there is a caveat: all fixed parameters must have the same value across the dataset in order to avoid creating a very large array whose size will scale with the number of different values of the non-free parameters.
Note
A good example of a linear model in the electron-microscopy field is an Energy-Dispersive
X-ray Spectroscopy (EDS) dataset, which typically consists of a polynomial background and
Gaussian peaks with well-defined energy (Gaussian.centre) and peak widths
(Gaussian.sigma). This dataset can be fit extremely fast with a linear optimizer.
There are two implementations of linear least squares fitting in hyperspy:
the
'lstsq'optimizer, which usesnumpy.linalg.lstsq(), ordask.array.linalg.lstsq()for lazy signals.the
'ridge_regression'optimizer, which supports regularization (seesklearn.linear_model.Ridgefor arguments to pass tofit()), but does not support lazy signals.
As for non-linear least squares fitting, weighted least squares is supported.
In the following example, we first generate a 300x300 navigation signal of varying total intensity,
and then populate it with an EDS spectrum at each point. The signal can be fitted with a polynomial
background and a Gaussian for each peak. Hyperspy automatically adds these to the model, and fixes
the centre and sigma parameters to known values. Fitting this model with a non-linear optimizer
can about half an hour on a decent workstation. With a linear optimizer, it takes seconds.
>>> nav = hs.signals.Signal2D(np.random.random((300, 300))).T
>>> s = hs.datasets.example_signals.EDS_SEM_Spectrum() * nav
>>> m = s.create_model()
>>> m.multifit(optimizer='lstsq')
Standard errors for the parameters are by default not calculated when the dataset
is fitted in vectorized fashion, because it has large memory requirement.
If errors are required, either pass calculate_errors=True as an argument
to multifit(), or rerun
multifit() with a nonlinear optimizer,
which should run fast since the parameters are already optimized.
None of the linear optimizers currently support bounds.
Optimization results
After fitting the model, details about the optimization
procedure, including whether it finished successfully,
are returned as scipy.optimize.OptimizeResult object,
according to the keyword argument return_info=True.
These details are often useful for diagnosing problems such
as a poorly-fitted model or a convergence failure.
You can also access the object as the fit_output attribute:
>>> m.fit()
<scipy.optimize.OptimizeResult object>
>>> type(m.fit_output)
<scipy.optimize.OptimizeResult object>
You can also print this information using the
print_info keyword argument:
# Print the info to stdout
>>> m.fit(optimizer="L-BFGS-B", print_info=True)
Fit info:
optimizer=L-BFGS-B
loss_function=ls
bounded=False
grad="fd"
Fit result:
hess_inv: <3x3 LbfgsInvHessProduct with dtype=float64>
message: b'CONVERGENCE: REL_REDUCTION_OF_F_<=_FACTR*EPSMCH'
nfev: 168
nit: 32
njev: 42
status: 0
success: True
x: array([ 9.97614503e+03, -1.10610734e-01, 1.98380701e+00])
Goodness of fit
The chi-squared, reduced chi-squared and the degrees of freedom are
computed automatically when fitting a (weighted) least-squares model
(i.e. only when loss_function="ls"). They are stored as signals, in the
chisq, red_chisq and
dof attributes of the model respectively.
Warning
Unless metadata.Signal.Noise_properties.variance contains
an accurate estimation of the variance of the data, the chi-squared and
reduced chi-squared will not be computed correctly. This is true for both
homocedastic and heteroscedastic noise.
Visualizing the model
To visualise the result use the plot() method:
>>> m.plot() # Visualise the results
By default only the full model line is displayed in the plot. In addition, it
is possible to display the individual components by calling
enable_plot_components() or directly using
plot():
>>> m.plot(plot_components=True) # Visualise the results
To disable this feature call
disable_plot_components().
New in version 1.4: Signal1D.plot keyword arguments
All extra keyword argments are passes to the plot() method of the
corresponing signal object. For example, the following plots the model signal
figure but not its navigator:
>>> m.plot(navigator=False)
By default the model plot is automatically updated when any parameter value
changes. It is possible to suspend this feature with
suspend_update().
Setting the initial parameters
Non-linear optimization often requires setting sensible starting parameters. This can be done by plotting the model and adjusting the parameters by hand.
Changed in version 1.3: All notebook_interaction() methods renamed to gui(). The
notebook_interaction() methods will be removed in 2.0
If running in a Jupyter Notebook, interactive widgets can be used to
conveniently adjust the parameter values by running
gui() for BaseModel,
Component and
Parameter.
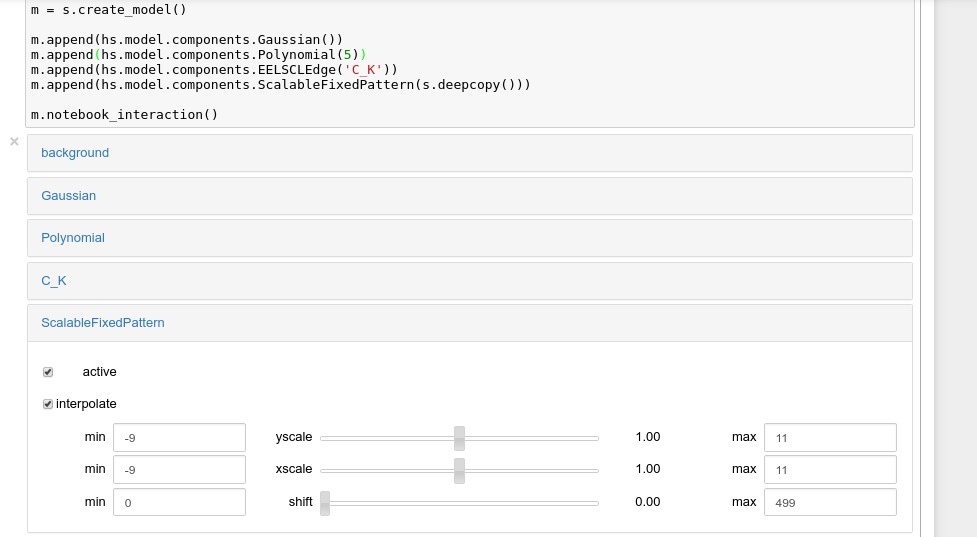
Interactive widgets for the full model in a Jupyter notebook. Drag the sliders to adjust current parameter values. Typing different minimum and maximum values changes the boundaries of the slider.
Also, enable_adjust_position() provides an
interactive way of setting the position of the components with a
well-defined position.
disable_adjust_position() disables the tool.
Interactive component position adjustment tool. Drag the vertical lines to set the initial value of the position parameter.
Exclude data from the fitting process
The following BaseModel methods can be used to exclude
undesired spectral channels from the fitting process:
In 2D models, those methods are not implemented and the
m.channel_switches attribute of a model can be set using boolean arrays of the
same shape as the data’s signal, where True means that the datapoint
will be used in the fitting routine.
The example below shows how a boolean array can be easily created from the
signal and how the isig syntax can be used to define the signal range.
>>> # Create a sample 2D gaussian dataset
>>> g = hs.model.components2D.Gaussian2D(
... A=1, centre_x=-5.0, centre_y=-5.0, sigma_x=1.0, sigma_y=2.0,)
>>> scale = 0.1
>>> x = np.arange(-10, 10, scale)
>>> y = np.arange(-10, 10, scale)
>>> X, Y = np.meshgrid(x, y)
>>> im = hs.signals.Signal2D(g.function(X, Y))
>>> im.axes_manager[0].scale = scale
>>> im.axes_manager[0].offset = -10
>>> im.axes_manager[1].scale = scale
>>> im.axes_manager[1].offset = -10
>>> m = im.create_model() # Model initialisation
>>> gt = hs.model.components2D.Gaussian2D()
>>> m.append(gt)
>>> # Create a boolean signal of the same shape as the signal space of im
>>> # and with all values set to `False`.
>>> signal_mask = hs.signals.Signal2D(np.zeros_like(im(), dtype=bool))
>>> # Specify the signal range using the isig syntax
>>> signal_mask.isig[-7.:-3.,-9.:-1.] = True
>>> m.channel_switches = signal_mask.data # Set channel switches
>>> m.fit()
Fitting multidimensional datasets
To fit the model to all the elements of a multidimensional dataset, use
multifit():
>>> m.multifit() # warning: this can be a lengthy process on large datasets
multifit() fits the model at the first position,
stores the result of the fit internally and move to the next position until
reaching the end of the dataset.
Note
Sometimes this method can fail, especially in the case of a TEM spectrum image of a particle surrounded by vacuum (since in that case the top-left pixel will typically be an empty signal).
To get sensible starting parameters, you can do a single
fit() after changing the active position
within the spectrum image (either using the plotting GUI or by directly
modifying s.axes_manager.indices as in Setting axis properties).
After doing this, you can initialize the model at every pixel to the
values from the single pixel fit using m.assign_current_values_to_all(),
and then use multifit() to perform the fit over
the entire spectrum image.
New in version 1.6: New optional fitting iteration path “serpentine”
Typically, curve fitting on a multidimensional dataset happens in the following
manner: Pixels are fit along the row from the first index in the first row, and once the
last pixel in the row is reached, one proceeds from the first index in the second row.
Since the fitting procedure typically uses the fit of the previous pixel
as the starting point for the next, a common problem with this fitting iteration
path is that the fitting fails going from the end of one row to the beginning of
the next, as the spectrum can change abruptly. This kind of iteration path is
the default in HyperSpy (but will change to 'serpentine' in HyperSpy version
2.0). It can be explicitly set using the multifit()
iterpath='flyback' argument.
A simple solution to the flyback fitting problem is to iterate through the
signal indices in a horizontal serpentine pattern, as seen on the image below.
This alternate iteration method can be enabled by the multifit()
iterpath='serpentine' argument. The serpentine pattern supports n-dimensional
navigation space, so the first index in the second frame of a three-dimensional
navigation space will be at the last position of the previous frame.
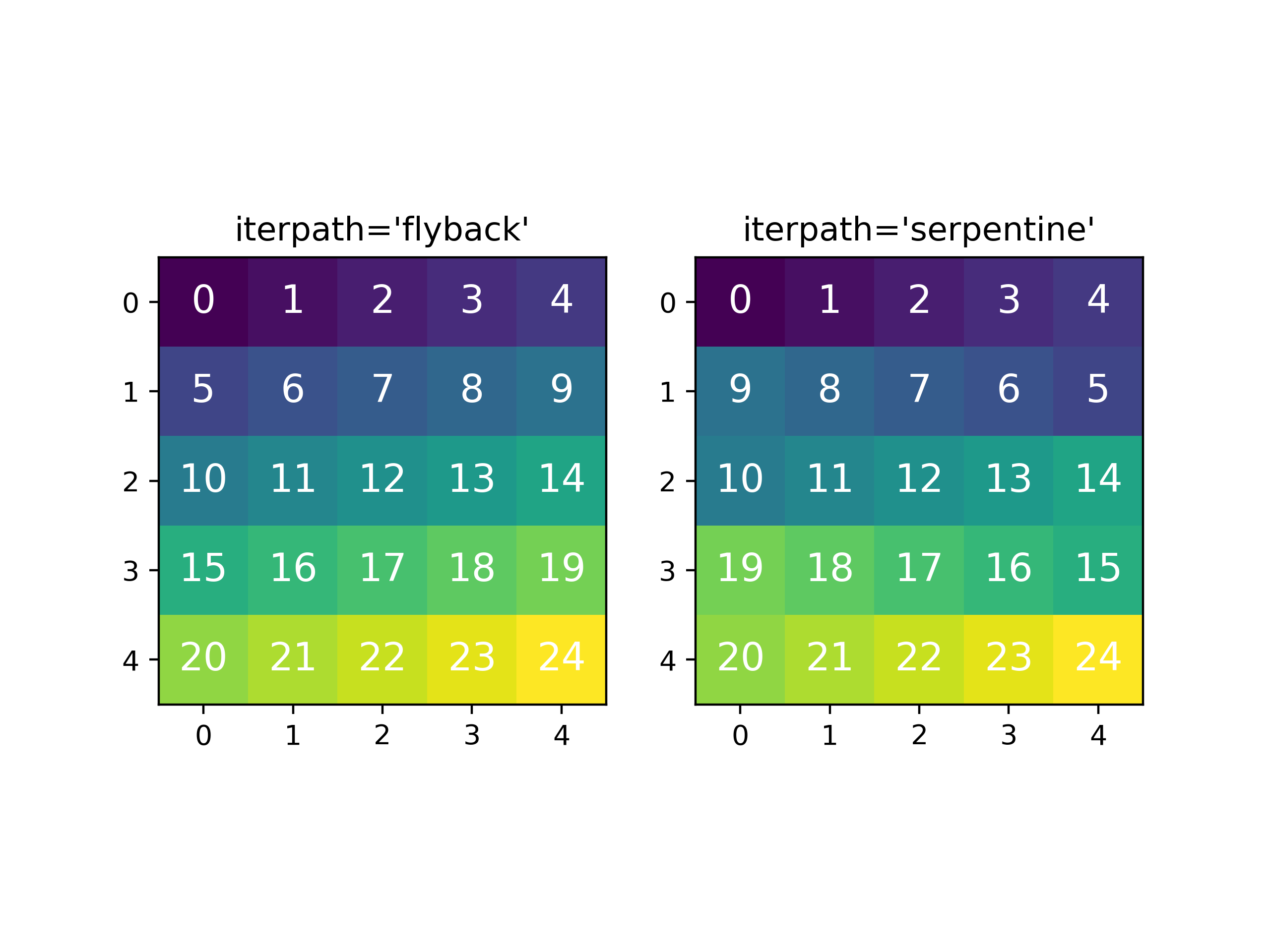
Comparing the scan patterns generated by the 'flyback' and 'serpentine'
iterpath options for a 2D navigation space. The pixel intensity and number refers
to the order that the signal is fitted in.
In addition to 'serpentine' and 'flyback', iterpath can take as argument any list
or array of indices, or a generator of such, as explained in the Iterating AxesManager section.
Sometimes one may like to store and fetch the value of the parameters at a
given position manually. This is possible using
store_current_values() and
fetch_stored_values().
Visualising the result of the fit
The BaseModel plot_results(),
Component plot() and
Parameter plot() methods
can be used to visualise the result of the fit when fitting multidimensional
datasets.
Storing models
Multiple models can be stored in the same signal. In particular, when
store() is called, a full “frozen” copy of the model
is stored in stored in the signal’s ModelManager,
which can be accessed in the models attribute (i.e. s.models)
The stored models can be recreated at any time by calling
restore() with the stored
model name as an argument. To remove a model from storage, simply call
remove().
The stored models can be either given a name, or assigned one automatically. The automatic naming follows alphabetical scheme, with the sequence being (a, b, …, z, aa, ab, …, az, ba, …).
Note
If you want to slice a model, you have to perform the operation on the model itself, not its stored version
Warning
Modifying a signal in-place (e.g. map(),
crop(),
align1D(),
align2D() and similar)
will invalidate all stored models. This is done intentionally.
Current stored models can be listed by calling s.models:
>>> m = s.create_model()
>>> m.append(hs.model.components1D.Lorentzian())
>>> m.store('myname')
>>> s.models
└── myname
├── components
│ └── Lorentzian
├── date = 2015-09-07 12:01:50
└── dimensions = (|100)
>>> m.append(hs.model.components1D.Exponential())
>>> m.store() # assign model name automatically
>>> s.models
├── a
│ ├── components
│ │ ├── Exponential
│ │ └── Lorentzian
│ ├── date = 2015-09-07 12:01:57
│ └── dimensions = (|100)
└── myname
├── components
│ └── Lorentzian
├── date = 2015-09-07 12:01:50
└── dimensions = (|100)
>>> m1 = s.models.restore('myname')
>>> m1.components
# | Attribute Name | Component Name | Component Type
---- | ------------------- | -------------------- | --------------------
0 | Lorentzian | Lorentzian | Lorentzian
Saving and loading the result of the fit
To save a model, a convenience function save() is
provided, which stores the current model into its signal and saves the
signal. As described in Storing models, more than just one
model can be saved with one signal.
>>> m = s.create_model()
>>> # analysis and fitting goes here
>>> m.save('my_filename', 'model_name')
>>> l = hs.load('my_filename.hspy')
>>> m = l.models.restore('model_name') # or l.models.model_name.restore()
For older versions of HyperSpy (before 0.9), the instructions were as follows:
Note that this method is known to be brittle i.e. there is no guarantee that a version of HyperSpy different from the one used to save the model will be able to load it successfully. Also, it is advisable not to use this method in combination with functions that alter the value of the parameters interactively (e.g. enable_adjust_position) as the modifications made by this functions are normally not stored in the IPython notebook or Python script.
To save a model:
Save the parameter arrays to a file using
save_parameters2file().Save all the commands that used to create the model to a file. This can be done in the form of an IPython notebook or a Python script.
(Optional) Comment out or delete the fitting commands (e.g.
multifit()).To recreate the model:
Execute the IPython notebook or Python script.
Use
load_parameters_from_file()to load back the parameter values and arrays.
Exporting the result of the fit
The BaseModel export_results(),
Component export() and
Parameter export()
methods can be used to export the result of the optimization in all supported
formats.
Batch setting of parameter attributes
The following model methods can be used to ease the task of setting some important parameter attributes. These can also be used on a per-component basis, by calling them on individual components.
Fitting big data
See the section in Binned and unbinned signals working with big data.
Smart Adaptive Multi-dimensional Fitting (SAMFire)
SAMFire (Smart Adaptive Multi-dimensional Fitting) is an algorithm created to reduce the starting value (or local / false minima) problem, which often arises when fitting multi-dimensional datasets.
The algorithm will be described in full when accompanying paper is published, but we are making the implementation available now, with additional details available in the following conference proceeding.
The idea
The main idea of SAMFire is to change two things compared to the traditional way of fitting datasets with many dimensions in the navigation space:
Pick a more sensible pixel fitting order.
Calculate the pixel starting parameters from already fitted parts of the dataset.
Both of these aspects are linked one to another and are represented by two different strategy families that SAMFfire uses while operating.
Strategies
During operation SAMFire uses a list of strategies to determine how to select the next pixel and estimate its starting parameters. Only one strategy is used at a time. Next strategy is chosen when no new pixels can be fitted with the current strategy. Once either the strategy list is exhausted or the full dataset fitted, the algorithm terminates.
There are two families of strategies. In each family there may be many strategies, using different statistical or significance measures.
As a rule of thumb, the first strategy in the list should always be from the local family, followed by a strategy from the global family.
Local strategy family
These strategies assume that locally neighbouring pixels are similar. As a result, the pixel fitting order seems to follow data-suggested order, and the starting values are computed from the surrounding already fitted pixels.
More information about the exact procedure will be available once the accompanying paper is published.
Global strategy family
Global strategies assume that the navigation coordinates of each pixel bear no relation to it’s signal (i.e. the location of pixels is meaningless). As a result, the pixels are selected at random to ensure uniform sampling of the navigation space.
A number of candidate starting values are computed form global statistical measures. These values are all attempted in order until a satisfactory result is found (not necessarily testing all available starting guesses). As a result, on average each pixel requires significantly more computations when compared to a local strategy.
More information about the exact procedure will be available once the accompanying paper is published.
Seed points
Due to the strategies using already fitted pixels to estimate the starting values, at least one pixel has to be fitted beforehand by the user.
The seed pixel(s) should be selected to require the most complex model present in the dataset, however in-built goodness of fit checks ensure that only sufficiently well fitted values are allowed to propagate.
If the dataset consists of regions (in the navigation space) of highly dissimilar pixels, often called “domain structures”, at least one seed pixel should be given for each unique region.
If the starting pixels were not optimal, only part of the dataset will be fitted. In such cases it is best to allow the algorithm terminate, then provide new (better) seed pixels by hand, and restart SAMFire. It will use the new seed together with the already computed parts of the data.
Usage
After creating a model and fitting suitable seed pixels, to fit the rest of the multi-dimensional dataset using SAMFire we must create a SAMFire instance as follows:
>>> samf = m.create_samfire(workers=None, ipyparallel=False)
By default SAMFire will look for an ipyparallel cluster for the
workers for around 30 seconds. If none is available, it will use
multiprocessing instead. However, if you are not planning to use ipyparallel,
it’s recommended specify it explicitly via the ipyparallel=False argument,
to use the fall-back option of multiprocessing.
By default a new SAMFire object already has two (and currently only) strategies
added to its strategies list:
>>> samf.strategies
A | # | Strategy
-- | ---- | -------------------------
x | 0 | Reduced chi squared strategy
| 1 | Histogram global strategy
The currently active strategy is marked by an ‘x’ in the first column.
If a new datapoint (i.e. pixel) is added manually, the “database” of the
currently active strategy has to be refreshed using the
refresh_database() call.
The current strategy “database” can be plotted using the
plot() method.
Whilst SAMFire is running, each pixel is checked by a goodness_test,
which is by default
red_chisq_test,
checking the reduced chi-squared to be in the bounds of [0, 2].
This tolerance can (and most likely should!) be changed appropriately for the data as follows:
>>> samf.metadata.goodness_test.tolerance = 0.3 # use a sensible value
The SAMFire managed multi-dimensional fit can be started using the
start() method. All keyword arguments are passed to
the underlying (i.e. usual) fit() call:
>>> samf.start(optimizer='lm', bounded=True)