Note
Go to the end to download the full example code.
Simple simulation (2 Gaussians)#
Creates a 2D hyperspectrum consisting of two Gaussians and plots it.
This example can serve as starting point to test other functionalities on the simulated hyperspectrum.
import numpy as np
import hyperspy.api as hs
import matplotlib.pyplot as plt
# Create an empty spectrum
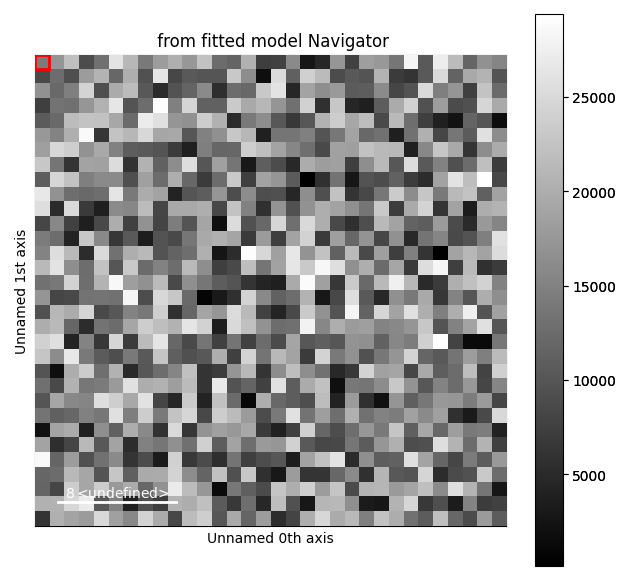
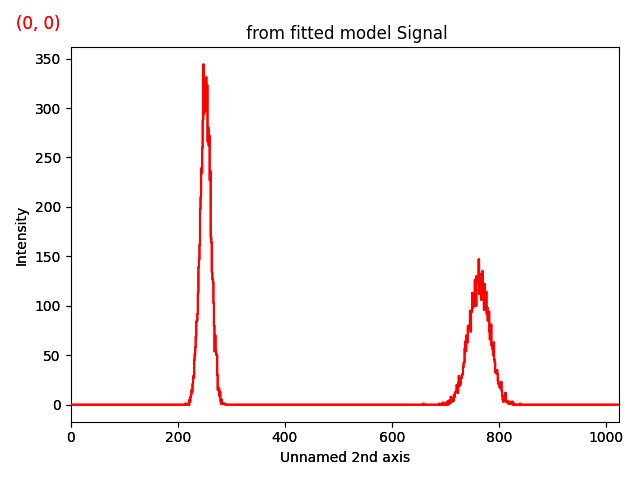
s = hs.signals.Signal1D(np.zeros((32, 32, 1024)))
# Generate some simple data: two Gaussians with random centers and area
# First we create a model
m = s.create_model()
# Define the first gaussian
gs1 = hs.model.components1D.Gaussian()
# Add it to the model
m.append(gs1)
# Set the parameters
m.set_parameters_value('sigma', 10, component_list=[gs1])
# Make the center vary in the -5,5 range around 128
gs1.centre.map['values'][:] = 256 + (np.random.random((32, 32)) - 0.5) * 10
gs1.centre.map['is_set'][:] = True
# Make the area vary between 0 and 10000
gs1.A.map['values'][:] = 10000 * np.random.random((32, 32))
gs1.A.map['is_set'][:] = True
# Second gaussian
gs2 = hs.model.components1D.Gaussian()
# Add it to the model
m.append(gs2)
# Set the parameters
m.set_parameters_value('sigma', 20, component_list=[gs2])
# Make the center vary in the -10,10 range around 768
gs2.centre.map['values'][:] = 768 + (np.random.random((32, 32)) - 0.5) * 20
gs2.centre.map['is_set'][:] = True
# Make the area vary between 0 and 20000
gs2.A.map['values'][:] = 20000 * np.random.random((32, 32))
gs2.A.map['is_set'][:] = True
# Create the dataset
s_model = m.as_signal()
# Add noise
s_model.set_signal_origin("simulation")
s_model.add_poissonian_noise()
# Plot the result
s_model.plot()
plt.show()
Total running time of the script: (0 minutes 0.880 seconds)